ProStaR
ProStaR, is devoted to the statistical analysis of label-free quantitative proteomics data produced by bottom-up LC-MS/MS experiments. Prostar project has two complementary aspects.
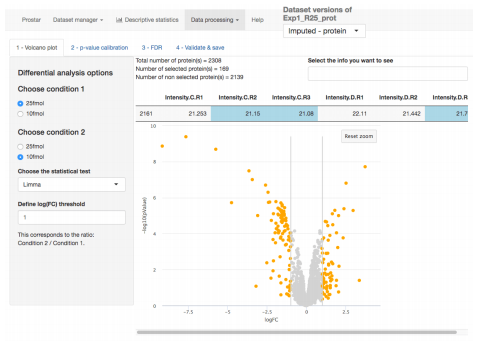
First, it gathers numerous independent methodological research works in biostatistics to improve the processing of label-free relative quantitative proteomics data. These improvements concerns both specific processing steps (such as missing value imputation, null hypothesis significance testing, false discovery rate control, etc.) and their organization into coherent pipelines [Wieczoreck et al., MiMB 2019; Jacob et al., Biostatistics 2018; Wieczoreck et al., Bioinformatics 2017; Burger, Journal of Proteome Research 2017; Giai Gianetto et al., Proteomics 2016; Lazar et al., Journal of Proteome Research 2016].
Second, ProStaR is a platform that is used on a daily basis by proteomics researchers and engineers to investigate their data. The software tool is built on R biostatistics routines (either pre-existing package or the aforementioned ProStaR research results) and web-based graphical user interfaces (Shiny technology). ProStaR is distributed through the BioConductor initiative, or via the project webpage www.prostar-proteomics.org. Over the last four years, Prostar have been downloaded in average 100 times/month (unique IPs).
Image credit: LittleAngell de Pixabay

