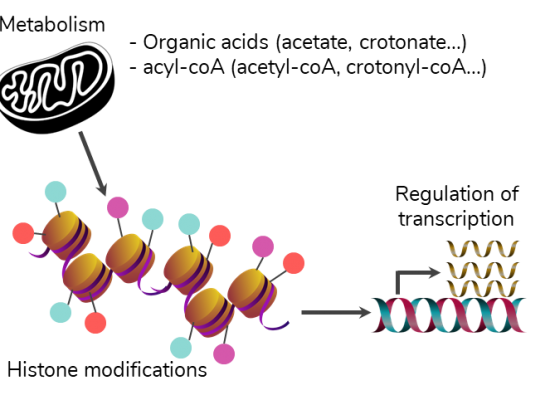
Histone modifications
Gene expression is largely regulated via the incorporation of histone variants and the dynamic modification of histones by various chemical groups. A tight regulation of these mechanisms is necessary during cellular differentiation processes; dysregulations are at stake in numerous diseases, including developmental defects, cancers and neurodegenerative diseases.
Tackling histone sequence variants by proteomics
Histone sequence variants, which may differ by just a handful of amino acids over the protein sequence, can be precisely studied by proteomic analysis. To confidently carry out such studies, we first established a database containing an exhaustive and non-redundant list of histone sequences for mouse and human [1]. In the context of mouse spermatogenesis that is characterized by very dynamic gene expression changes and dramatic chromatin remodeling, we demonstrated the feasibility of identifying and quantifying numerous variants of histones H2A and H2B, including sequences that differ by a single amino acid [2]. Recently, we assessed the existence at the protein level of mouse-specific variants of histone H3 in mouse testis and brain [3]. This work allowed describing with great care the particular difficulty encountered when analyzing histones, which yield proteolytic peptides that might correspond to incredibly numerous combinations of variant x post-translational modifications (PTMs) of very same mass. This leads to challenging proteomic analyses that require both mastering the biochemical processing of histone samples before their analysis, and paying attention to low-mass informative features in fragmentation spectra [3,4,5,6].
Studying the effects on gene expression of the wealth of histone lysine post-translational modifications by multi-omics
Our research activities particularly aim at studying the effects on gene expression of the multiple PTMs that target histone lysine residues. In particular, various acylations have been described since 2007, which resemble acetylation, yet differ in length, hydrophobicity and charge, and emerge as being endowed with specific functions compared to acetylation. All these acylations connect the histone PTM profile to cellular metabolites, more precisely to acyls-CoA (e.g. acetyl-CoA, crotonyl-CoA, etc.) and to the corresponding carboxylates (acetate, crotonate, etc.). We have carried out such explorations in the context of mouse spermatogenesis [7,9]. In particular, the proteomic analysis of histones extracted from three cellular stages of mouse spermatogenesis revealed that crotonylation was of similar stoichiometry to acetylation on histone H3 Lysine 27 (H3K27cr). Epigenomic analyses of H3K27cr and of the canonical mark H3K27ac indicated both synergistic and specific actions of each histone modification at promoters and distal enhancers [7]. We are pursuing our efforts to characterize further acylations in mouse spermatogenesis, including the more recently described lactylation.
Over the last years, we have expanded the study of histone lysine acylations to pathologies in which epigenetic landscapes are rewired. In particular, we contribute to deciphering the links between dysregulated metabolism and revisited histone PTM profiles in Huntington’s disease (HD), a rare genetic neurodegenerative disease that primarily affects the striatum. To achieve this goal, we have established isotope tracing which allows following the kinetics of histone acetylation at multiple lysine residues after injection into mice of a heavy labeled metabolite such as glucose.
Over the next years, we wish to reinforce this research axis aiming at deciphering the links between imbalanced metabolism and epigenetic dysregulations, in particular in neurodegenerative diseases.
EDyP project leader
Delphine Pflieger
CNRS Researcher
Publications
- El Kennani S, Adrait S, Shaytan A, Khochbin S, Bruley C, Panchenko AR, Landsman D, Pflieger D, Govin J. MS_HistoneDB, a manually curated resource for proteomic analysis of human and mouse histones. Epigenetics & Chromatin, 2017, Jan 10;10:2. doi: 10.1186/s13072-016-0109-x.
- El Kennani S, Adrait A, Permiakova O, Hesse AM, Ialy-Radio C, Ferro M, Brun V, Cocquet J, Govin J, Pflieger D. Systematic quantitative analysis of H2A and H2B variants by targeted proteomics.Epigenetics & Chromatin, 2018, 2018 Jan 12;11(1):2. doi: 10.1186/s13072-017-0172-y.
- Hijazi H, Manessier J, Brugiere S, Ravnsborg T, Courçon M, Brule B, Merienne K, Jensen ON, Hesse AM, Bruley C, Pflieger D. Mind Your Spectra: Points to be Aware of When Validating the Identification of Isobaric Histone Peptidoforms. Proteome Res. 2025 May 2;24(5):2408-2418. doi: 10.1021/acs.jproteome.4c01056.
- El Kennani S, Crespo M, Govin J, Pflieger D. Proteomic analysis of histone variants and their PTMs: strategies and pitfalls. Proteomes, 2018, 2018 Jun 21;6(3):29. doi: 10.3390/proteomes6030029.
- Hseiky A, Crespo M, Kieffer-Jaquinod S, Fenaille F, Pflieger D.Small Mass but Strong Information: Diagnostic Ions Provide Crucial Clues to Correctly Identify Histone Lysine Modifications. Proteomes. 2021 Apr 23;9(2):18. doi: 10.3390/proteomes9020018.
- Geshkovski V, Hijazi H, Manessier J, Brugière S, Courçon M, Vachon G, Pflieger D, Carles CC. Quantitative Profiling of Histone Variants and Posttranslational Modifications by Tandem Mass Spectrometry in Arabidopsis. Methods Mol. Biol. 2025;2873:19-38. doi: 10.1007/978-1-0716-4228-3_2.
- Marion Crespo, Annelaure Damont, Melina Blanco, Emmanuelle Lastrucci, Sara El Kennani, Côme Ialy-Radio, Laila El Khattabi, Samuel Terrier, Mathilde Louwagie, Sylvie Kieffer-Jaquinod, Anne-Marie Hesse, Christophe Bruley, Sophie Chantalat, Jérôme Govin, François Fenaille, Christophe Battail, Julie Cocquet, Delphine Pflieger. Multi-omic analysis of gametogenesis reveals a novel signature at the promoters and distal enhancers of active genes.Nucleic Acids Res. 2020 May 7;48(8):4115-4138. doi: 10.1093/nar/gkaa163.
- Marion Crespo, Lacey J. Luense, Marie Arlotto, Jialei Hu, Jean Dorsey, Encar García-Oliver, Parisha P. Shah, Delphine Pflieger, Shelley L. Berger, Jerome Govin. Systematic genetic and proteomic screens during gametogenesis identify H2BK34 methylation as an evolutionary conserved meiotic mark. Epigenetics & Chromatin, 2020 Sep 15;13(1):35. doi: 10.1186/s13072-020-00349-5.
- Blanco M, El Khattabi L, Gobé C, Crespo M, Coulée M, de la Iglesia A, Ialy-Radio C, Lapoujade C, Givelet M, Delessard M, Seller-Corona I, Yamaguchi K, Vernet N, Van Leeuwen F, Lermine A, Okada Y, Daveau R, Oliva R, Fouchet P, Ziyyat A, Pflieger D, Cocquet J. DOT1L regulates chromatin reorganization and gene expression during sperm differentiation. EMBO Rep. 2023 Jun 5;24(6):e56316. doi: 10.15252/embr.202256316. Epub 2023 Apr 26.
Fundings
- PhD fellowship from Association Huntington France to Hisham Altoufaily (2025-2027, co-direction Delphine Pflieger and Karine Merienne): “Study of the links between the dysregulations of metabolism and histone post-translational modifications in Huntington’s disease”
- ANR-funded project CRE-Subnucleosome2 (2023-2027, coordinator Matthieu Gérard): “Physical and functional characterization of a new class of subnucleosomal particles located at cis-regulatory elements in mammals”
- ANR-funded project HD-EPIeNERGY (2022-2026, coordinator Karine Merienne): “Linking energy metabolism to chromatin in Huntington’s disease”
- ANR-funded project CHROMACYL (2022-2026, coordinator Delphine Pflieger, partners Julie Cocquet and Christophe Battail), including the PhD of Julie Manessier: “Studying novel histone acylations in the regulation of gene expression”
- PhD fellowship to Olivier Feudjio (CIFRE, Adlin Science, 2022-2025, co-direction Delphine Pflieger and Julie Cocquet): “Investigation of the regulation of gene expression via alternative splicing and the interplay with epigenetic modifications in the context of mouse spermatogenesis”.
- GRAL PhD fellowship to Hassan Hijazi (2021-2024, co-direction Delphine Pflieger and Karine Merienne): “Altered acylation dynamics of histone H3 Lysine 27 (H3K27): a mechanism contributing to transcriptional deregulation in Huntington’s disease?”
- UGA PhD fellowship to Marion Crespo (2016-2019)
- CEA PhD fellowship to Sara El Kennani (2015-2019)
