Author: EDyP Lab
NEMS
This project aimed to push back the limits of mass range detection using Nano-Electro-Mechanical resonators (NEMS). NEMS were developed for various applications […]
Proline 2.0 Release
Proline Release 2.0 New datastore architecture, separation of Instrument and Fractionnation rules, add new export formats, enhancement of PTM Sites managment and […]
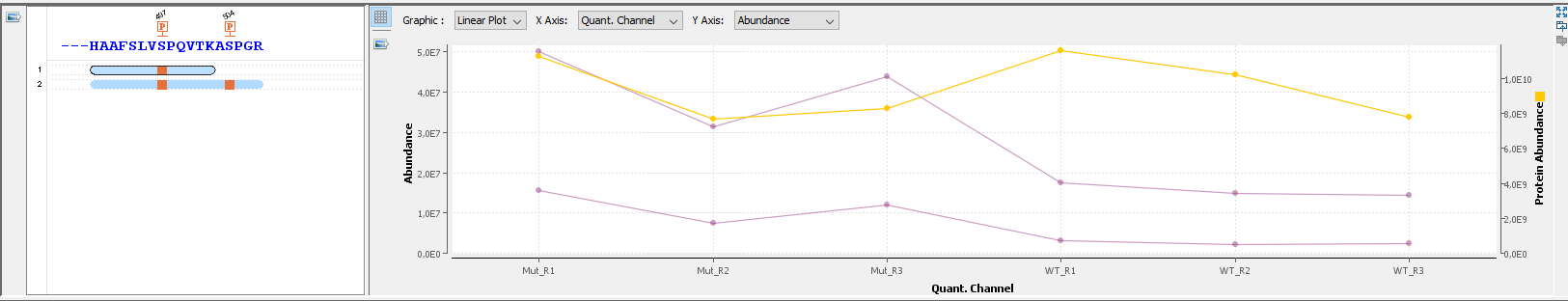
Proline
Proline Proline is a landmark software suite for quantitative proteomics allowing to collect, process, visualize and publish proteomics datasets [Bouyssié et al., […]
ProStaR
ProStaR ProStaR, is devoted to the statistical analysis of label-free quantitative proteomics data produced by bottom-up LC-MS/MS experiments. Prostar project has two […]
ProteoRE
ProteoRE The development of ProteoRE (Proteomics Research Environment) is an online research service to assist biologists/clinicians in the interpretation of their “Omics” […]
Integrative Omics
Integrative omics to study gene expression regulation Gene expression is regulated in part by the incorporation of histone variants and by the […]
Plasma Biomarkers
Plasma biomarkers MS-based proteomics is particularly well-suited for identifying specifically and measuring accurately the abundance of protein biomarker candidates. During the last […]