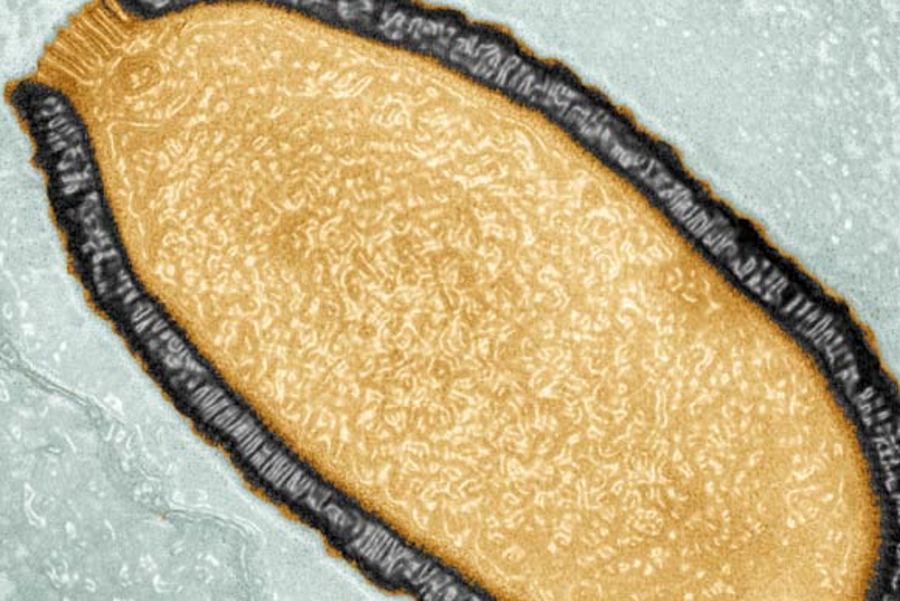
Functional and molecular characterization of giant viruses
The recent discovery in various spots around the world of novel giant viruses largely made of proteins with no homologs in other species raised many questions about their origin but also about the way they can manipulate the standard biochemical building blocks of the living world using vastly different proteins. We used various proteomic approaches to:
- characterize the protein content of different giant viruses. We notably identify the protein contents as well as the surfomes of different viruses using dedicated strategies. We implemented a label-free quantification strategy in order to estimate the protein stoichiometries within particles. Finally we characterized post-translational modifications carried by virion proteins.
- explore the infectious cycles of selected giant viruses. A label-free quantitative proteomic strategy was deployed to follow the relative abundances of viral and host proteins along the infectious cycle, highlighting notably the expression kinetics of viral proteins and the global preservation of the host cell proteome.
Collaborators
Laboratoire Information Génomique et Structurale, Marseille (https://www.igs.cnrs-mrs.fr/, Head : Chantal Abergel)
EDyP project leader
Publications
- Legendre, M., Alempic, J.-M., Philippe, N., Lartigue, A., Jeudy, S., Poirot, O., … Claverie, J.-M. (2019). Pandoravirus Celtis Illustrates the Microevolution Processes at Work in the Giant Pandoraviridae Genomes. Frontiers in Microbiology, 10, 430. https://doi.org/10.3389/fmicb.2019.00430
- Legendre, M., Fabre, E., Poirot, O., Jeudy, S., Lartigue, A., Alempic, J.-M., … Claverie, J.-M. (2018). Diversity and evolution of the emerging Pandoraviridae family. Nature Communications, 9(1), 2285. https://doi.org/10.1038/s41467-018-04698-4
- Fabre, E., Jeudy, S., Santini, S., Legendre, M., Trauchessec, M., Couté, Y., … Abergel, C. (2017). Noumeavirus replication relies on a transient remote control of the host nucleus. Nature Communications, 8, 15087. https://doi.org/10.1038/ncomms15087
- Legendre, M., Lartigue, A., Bertaux, L., Jeudy, S., Bartoli, J., Lescot, M., … Claverie, J.-M. (2015). In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proceedings of the National Academy of Sciences of the United States of America, 112(38), E5327-5335. https://doi.org/10.1073/pnas.1510795112
- Legendre, M., Bartoli, J., Shmakova, L., Jeudy, S., Labadie, K., Adrait, A., … Claverie, J.-M. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proceedings of the National Academy of Sciences of the United States of America, 111(11), 4274–4279. https://doi.org/10.1073/pnas.1320670111
- Philippe, N., Legendre, M., Doutre, G., Couté, Y., Poirot, O., Lescot, M., … Abergel, C. (2013). Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science (New York, N.Y.), 341(6143), 281–286. https://doi.org/10.1126/science.1239181
Funding
Image credit: Julia Bartoli / Chantal Abergel / IGS / CNRS / AMU.